Unveiling New Horizons in Glioma Research: The nSEA Algorithm
In the ever-evolving field of cancer research, breakthroughs that offer new insights into tumor classification and treatment are invaluable. Our recent study, “nSEA: n-Node Subnetwork Enumeration Algorithm Identifies Lower Grade Glioma Subtypes with Altered Subnetworks and Distinct Prognostics,” introduces a groundbreaking approach to understanding lower-grade gliomas (LGG) (Zhang et al., 2024). Here, I will talk about top findings of our study, highlighting its potential to improve glioma treatment and its clinical relevance.
A Novel LGG Subgroup
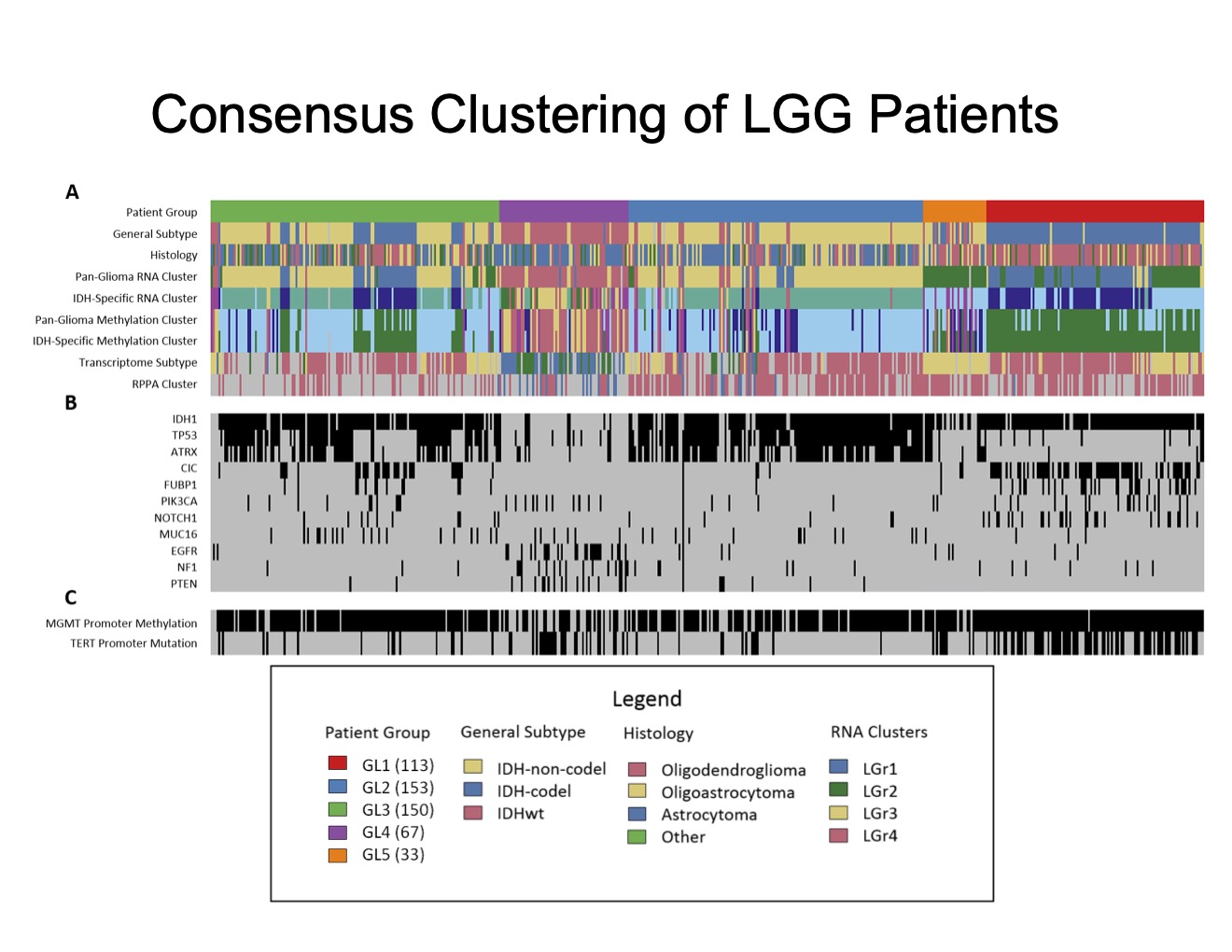
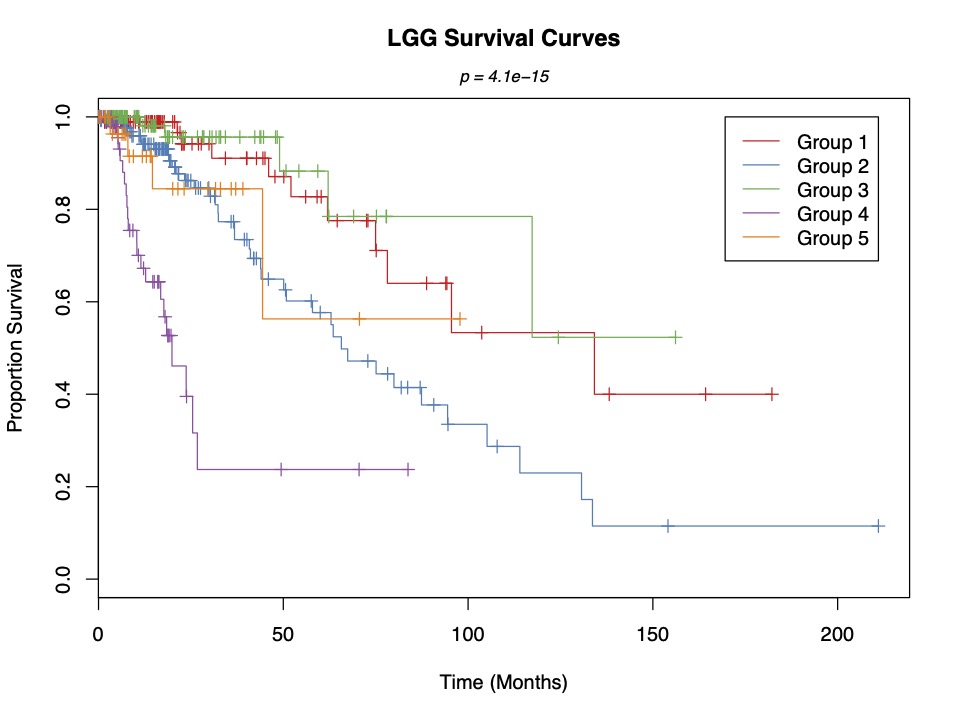
One of the most significant discoveries from our research is the identification of a previously unrecognized LGG patient subgroup. Using the nSEA algorithm, we analyzed gene expression profiles from 516 patients (TCGA) and a protein-protein interaction network, generating an astounding 25 million subnetworks. Remarkably, we distilled the initial 25 million subnetworks down to 92 key subnetworks using our nSEA algorithm. Through our unsupervised bottom-up approach we created classification of LGG patients into five distinct groups. Among these, one subgroup stood out due to its unique clinical features and subnetwork states, which had not been identified in prior studies.
This new subgroup lacks mutations in key genes such as EGFR, NF1, and PTEN, which are commonly associated with gliomas. Patients in this group exhibit distinct survival traits, suggesting that they may benefit from different therapeutic strategies compared to other LGG patients. This finding underscores the importance of molecular classification in providing personalized treatment plans and improving patient outcomes.
Transforming Treatment and Clinical Relevance
The nSEA algorithm’s ability to classify LGG patients into distinct subgroups based on molecular pathways has the potential to transform glioma treatment. By identifying patient groups with specific molecular alterations, clinicians can tailor treatment plans to target these unique features, potentially improving patient outcomes. For instance, the newly identified subgroup with a lack of EGFR, NF1, and PTEN mutations may respond better to therapies that are ineffective for other LGG patients.
Moreover, this study paves the way for further investigations into the mechanistic underpinnings of LGG subtypes. Understanding the biological processes driving these subgroups can lead to the development of new therapeutic targets and personalized interventions, ultimately advancing the field of glioma research.
Tools and Resources
The source code and supplementary data are accessible on GitHub.
Conclusion
The nSEA algorithm not only identifies a novel LGG subgroup but also provides a robust framework for future research and clinical applications. Engaging with this study can contribute to the growing body of knowledge that aims to improve glioma prognosis and treatment.
For those interested in exploring the full potential of the nSEA algorithm, the source code and supplementary data are readily available on GitHub.
References
2024
-
nSEA: n-Node Subnetwork Enumeration Algorithm Identifies Lower Grade Glioma Subtypes with Altered Subnetworks and Distinct PrognosticsIn PACIFIC SYMPOSIUM ON BIOCOMPUTING 2024 , 2024