Distinct Proteomic Signatures in Rapidly Progressive Alzheimer's Disease
We are pleased to announce the publication of our recent study investigating the molecular distinctions of rapidly progressive Alzheimer’s disease (rpAD). This work focuses on proteomic analyses of matched brain and cerebrospinal fluid (CSF) samples to identify biological features unique to rpAD, a rare and understudied form of Alzheimer’s disease.
Key findings
-
Integrated brain and CSF proteomics: Using fresh frozen tissue (FFT) from the medial temporal lobe and matched CSF samples from the same individuals, we identified a shared set of 35 proteins, many of which are associated with immune function and proteostasis.
-
Enhanced proteomic coverage with FFT: Compared to previous studies using formalin-fixed, paraffin-embedded (FFPE) tissue, FFT provided more comprehensive protein detection and highlighted additional biological pathways, particularly those related to neuroinflammation.
-
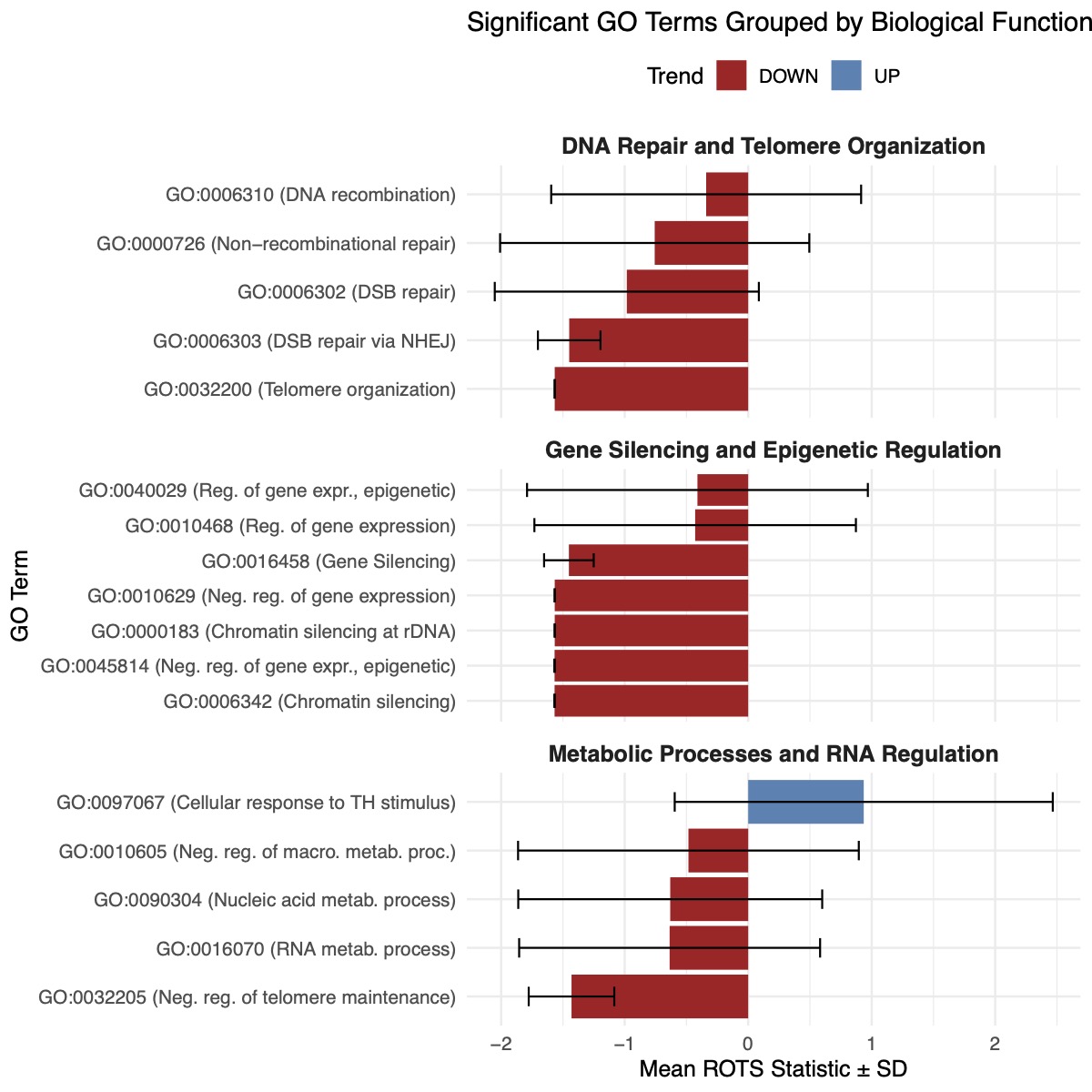
Distinct biological processes in rpAD: Differential expression and enrichment analyses revealed that rpAD is characterized by disruptions in DNA repair, epigenetic regulation, and extracellular matrix organization. These alterations suggest a systemic failure of cellular maintenance mechanisms rather than the selective upregulation of individual pathological processes.
-
Potential biomarkers for rapid progression: Five proteins—PGAM1, YWHAG, DLD, PARK7, and EPDR1—were consistently upregulated in both brain and CSF of rpAD cases, suggesting their potential utility as biomarkers for identifying rapidly progressive forms of AD.
-
Association with tau pathology: A coordinated downregulation of the shared protein signature was associated with lower tau (MAPT) levels in CSF, underscoring the relevance of this proteomic axis to core AD pathology.
This study supports the view that rpAD represents a distinct molecular subtype of Alzheimer’s disease. The findings may inform future efforts to develop diagnostic tools or therapeutic strategies tailored to this aggressive clinical phenotype.
Read the full paper: [coming soon](Bebek et al., 2025)
References
2025
-
Protein co-aggregates of dense core amyloid plaques and CSF differ in rapidly progressive Alzheimer’s disease and slower sporadic Alzheimer’s diseaseAlzheimer’s Research & Therapy., 2025